Simulating an oncology preclinical study with only 27 colon cancer patients derived PDX
Overview
Identifying the right patients who can benefit from a specific treatment or combination of treatment remains a challenging task, only becoming more difficult as the number of accessible therapies and their combinations increase. Patient derived xenograft mouse models (PDX) experiments combined with AI provide an opportunity to simulate a preclinical assessment using multiple mice. The platform was tested on colon cancer patients derived PDX.
Impact
• KEM® eXp.AI successfully simulated a preclinical study and identified biomarkers of drug efficacy and synergy.
• Such insights pave the way for innovative precision medicine clinical trials in which experiments performed in PDX and analysed using AI will deliver actionable hypothesis for patients’ inclusion and study design.
• The platform can be used for drug repositioning or identification of innovative drug combinations
Objectives
• Combine the power of PDX models with KEM® eXp.AI data analytics to simulate an oncology preclinical assessment.
• Systematically identify biomarkers of response predicting therapeutic efficacy, using state-of-the-art PDX models.
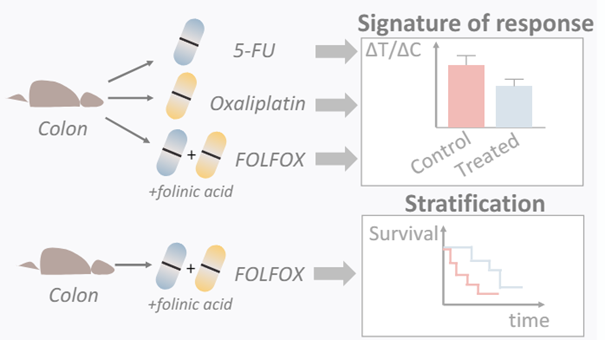

Method
The platform was tested on colon cancer patient derived PDX.
This study engaged an experimental setup involving 27 colon cancer PDX models, analyzing DNA and RNA to determine molecular characteristics linked to positive response to specific drug combinations. Survival rates were measured in PDXs treated with FOLFOX or placebo, providing a comparative view of treatment efficacy.
Results
• Comprehensive genomic analysis led to the identification of 24 candidate genes that may predict a positive response to treatment, including notable genes like TP53, NOTCH2, and ERBB2.
• Survival analysis of these genes in PDX models indicated a distinct subgroup of patients who might benefit from targeted treatment approaches, which could be crucial for future clinical applications.
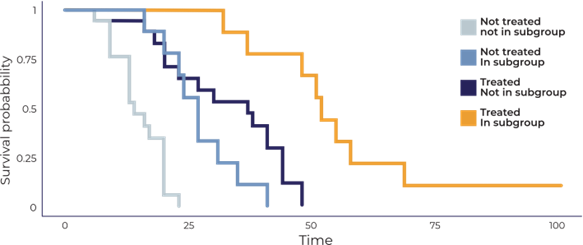

Survival analysis for each gene identified
