Oncology: Prostate cancer: KEM®-based genomic biomarker signature predicts neoadjuvant androgen deprivation therapy (NADT) response
Overview
This case study presents a novel Explainable AI-driven genomic signature, developed by Ariana Pharmaceuticals in collaboration with Philips, that predicts patient response to neoadjuvant androgen deprivation therapy (NADT) in localized high-risk prostate cancer.
The study was presented at ESMO 2025 (AI-Broadcast Session, Berlin) and marks an important milestone in the integration of AI-based precision medicine into oncology practice.
Using the KEM® Explainable AI platform, genomic and phenotypic data from 58 patients across two major clinical studies NCT02430480 (Wilkinson et al., 2021) and NCT02268175 (Tewari et al., 2021) were analyzed to develop and validate a predictive genomic signature distinguishing exceptional responders from poor responders to NADT.
Impact
-
- ● Identification of a novel explainable genomic signature capable of predicting NADT response in localized high-risk prostate cancer. (ICD-10 K71).
- ● The KEM® Biomarker signature provides interpretable patient-level predictions, paving the way for explainable AI in clinical oncology.
- ● The study demonstrates the feasibility of leveraging Explainable AI to stratify patients, improve therapy decision-making, and support future precision medicine applications.
- ● • Lays the groundwork for prospective validation and potential integration into clinical workflows.
Objectives:
-
- ● Develop a genomic predictive signature for prostate cancer prognosis in patients treated with NADT. (ICD-10 K71).
- ● Identify molecular features distinguishing exceptional responders from poor responders..
- ● The study demonstrates the feasibility of leveraging Explainable AI to stratify patients, improve therapy decision-making, and support future precision medicine applications.
- ● Validate and interpret predictive rules generated through the KEM® platform..
Method:
-
A curated list of 100 gene symbols was preselected based on biological relevance, including pathways related to:
-
- ● Viral-bacterial defense mechanisms
- ● T-cell receptor (TCR) signaling.
- ● Correlation with PDE4D7 expression (a known marker of androgen response)
Study Design:
Results:
- ● The best-performing KEM® Biomarker signature identified GUCY1A1, PLS3, ARSD, and CUX2 as key high-expression genes predictive of NADT response.
- ● The coverage matrix provided an explainable visualization of rule-based predictions at the patient level, allowing clinicians to interpret why a particular patient was predicted as a responder or non-responder.
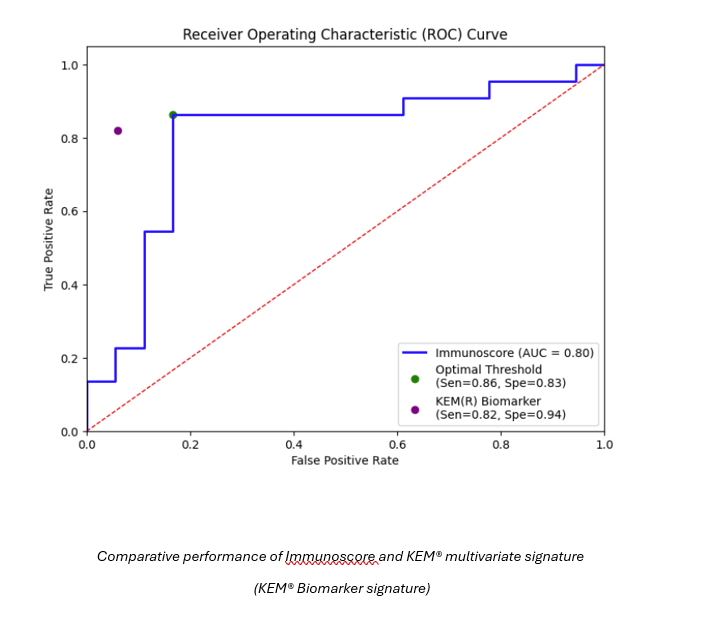
- ● Comparative performance against Immunoscore and traditional multivariate approaches demonstrated improved predictive power and transparency with the KEM® model.
- ● This multi-cohort collaboration with Philips confirms the potential of explainable AI in oncology biomarker development.