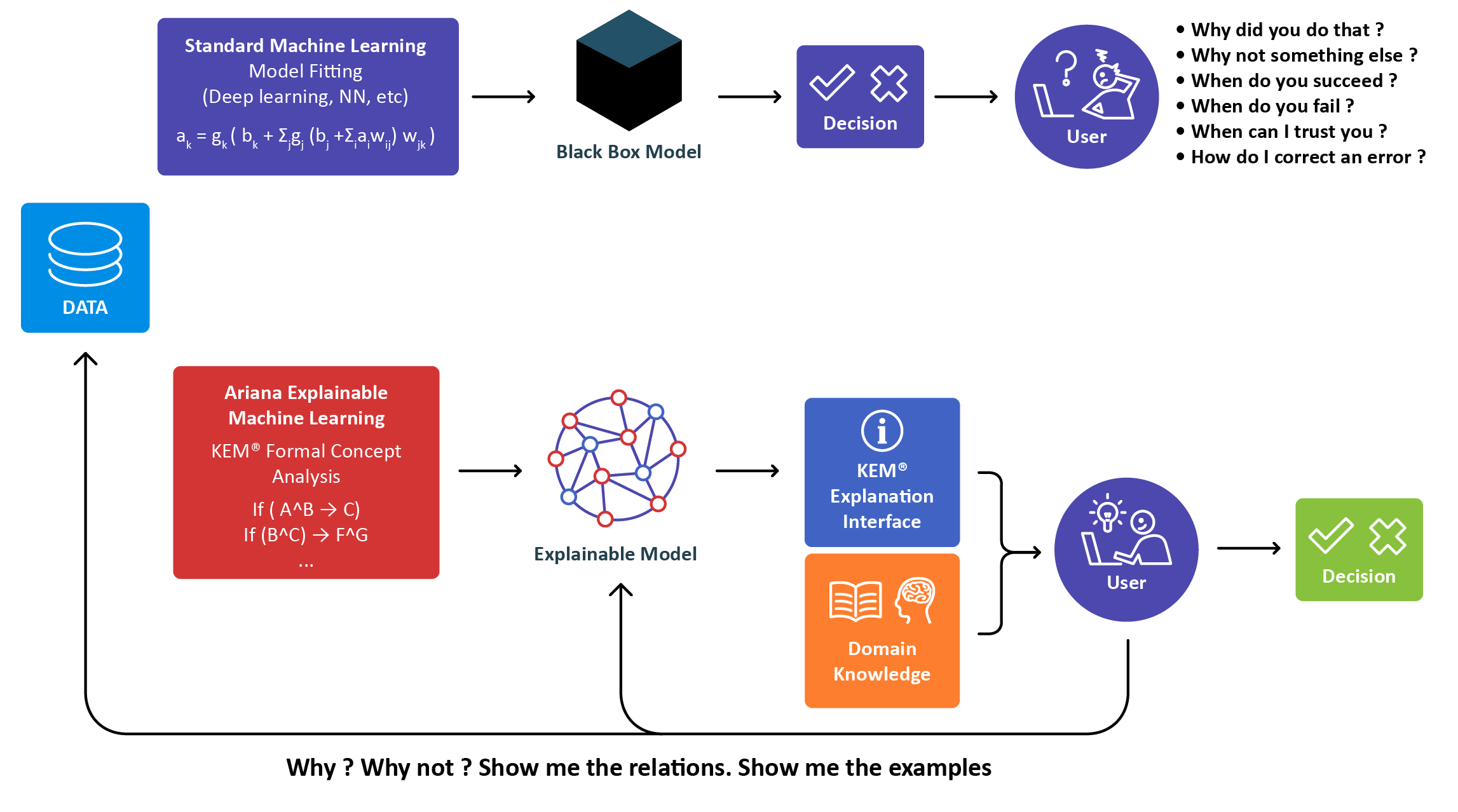
Our Technology
The next stage of human machine collaboration AI
which learns and explains why

KEM® is the result of over 15 years of development for the life sciences at Ariana by experts in Machine Learning, Statistics, Biology, biomarker research and drug development.
Our eXp.AI technology uncovers hidden signals and complex relationships that conventional statistical analyses
miss. It’s a unique approach that is both complementary to and supported by statistical analyses, guaranteeing
robust, reproducible results. We can easily analyze complex combinations of data including clinical, imaging,
metabolic, biochemical and NGS/omics.
Using existing and new data provided for your clinical trial, Ariana combines baseline characteristics, gene expression, protein measurements, matebolimic date. We generate all logical signatures, evaluate each to deliver signatures you can count on and ranks each combination of these data points showing you the most
important to your trail.
“Ariana®: The Explainable Artificial Intelligence Driven Precision Medicine Company “
With KEM®, the signatures are easily interpretable
The KEM® approach identifies all possible signatures. The ranking and filtering are performed explicitly based on classification performance, signature simplicity (length), as well as any additional criteria such as biological knowledge (pathways enrichment) or clinical relevance (targeting specific patient subgroups) etc.
“Al Driven Knowledge Integration and Discovery Platform Aiming at Regulatory Strategy”
KEM® can identify all relevant relationships between variables
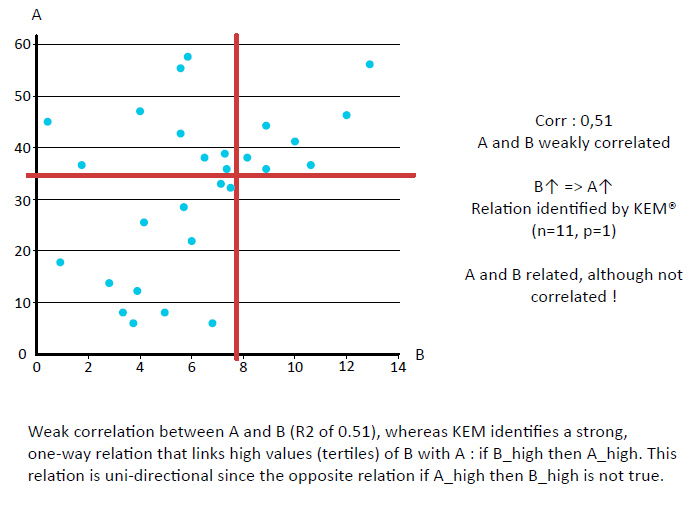

KEM® delivers unsupervised, unbiased, total data exploration.
This avoids many of the over-fitting issues, since there is no arbitrary optimization.
This can be compared with Random Forest, for example, where you may get rules that indicate that BMI should be < 17.003, which is unlikely to hold any medical meaning.
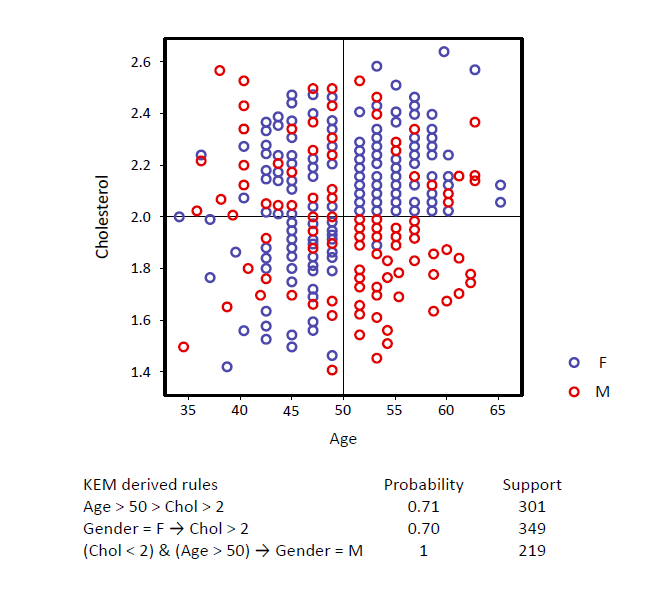

KEM® generates compact, interpretable signatures
with high specificity and sensitivity
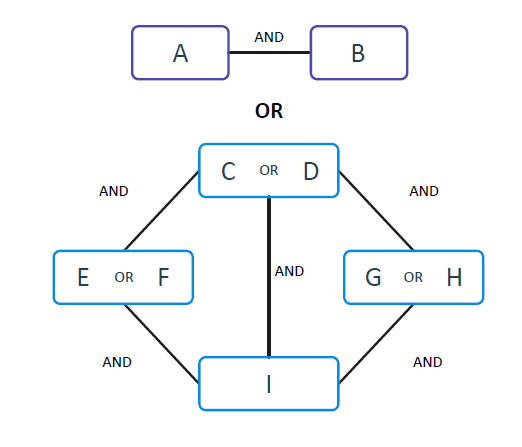

They contribute to the increase of the specificity of the signature. Multiple ANDs are then combined through OR clauses, increasing sensitivity. In the FCA paradigm, there is no need for global convergence; hence the system can identify multiple local minima that are connected via ORs, enabling the effective and efficient analysis of heterogeneous data. The signatures identified by KEM® are comprised of a non-linear combination of AND and OR functions. The result is usually a much more compact signature
Latest Scientific Publications
Clinical Trials on Alzheimer's Disease
Conference (CTAD) 2020
” ANAVEX®2-73 (Blarcamesine) Currently in Phase 2b/3 Early)...
Pharmaceutical Users Software Exchange US 2020
“Artificial Intelligence ( AI ) Enhanced Precision Medicine Drug Development”
Clinical Trials on Alzheimer's Disease conference (CTAD) 2019, San Diego, USA
“Novel analytics framework for...
Clinical Trials on Alzheimer's Disease conference (CTAD) 2018, Barcelona, Spain
“Longitudinal 148-Week.
